Loading NetCDFs in TensorFlow
NetCDF is a ubiquitous binary data format in the geosciences. While it may not be widely known in other domains it has several advantages. Most importantly it can efficiently store large amounts of data and is self-describing. In my opinion, NetCDF is as close to plain text as binary data can get in terms of universality. Seeing the contents of a dataset on a high level requires a simple call to a CLI ncdump -h. This makes it a perfect format for sharing climate simulation outputs, weather data, etc.
Unfortunately, TensorFlow does not natively support NetCDF. So what are we to do?
In this post, I benchmark several ways of loading a dataset from NetCDF. The goal is not to demonstrate a feature-complete solution, but to introduce a range of methods and rigorously compare their performance. Perhaps surprisingly, I show that NetCDF is a perfectly acceptable format for training ML models and may outperform some “native” TensorFlow formats.
Benchmarks
Geoscientists often start a machine learning project with a folder of NetCDFs, so I will benchmark a similar task: load a folder of NetCDFs into tf.data.Dataset object and iterate through all the samples to mimic one epoch of a machine learning training loop.
For simplicity, each NetCDF file contains a single variable “a”.
For a dataset of a given size, a key question is how many files should we divide it into to achieve good performance. If the file size is too large (e.g. multiple GB) then it can be unwieldy to inspect and experiment with especially if it is stored in cloud storage. OTOH, opening up many 1000s of tiny files is sure to bog down any ML training.
To set up these experiments, save_dirs is a function that divides an artificial dataset of a certain size into many NetCDFs of a certain size. Each NetCDF contains a vector of random values “a”.
import numpy
import netCDF4 as nc
import xarray
import tensorflow_datasets as tfds
import tensorflow as tf
import os
import shutil
import glob
import plotly.express as px
print("Tensorflow:", tf.version.VERSION)
print("Xarray:", xarray.__version__)
print("netCDF4:", nc.__version__)
# initialize directory of files
dir_ = "ncfiles"
kb = 1_024
mb = kb * kb
def save_dirs(target_size: int, total_size: int, dir_: str):
shutil.rmtree(dir_, ignore_errors=True)
os.makedirs(dir_)
nfiles = int(total_size / target_size)
n = int(target_size / 8)
ds = xarray.DataArray(numpy.random.uniform(size=(n // 64, 64))).to_dataset(name="a")
os.makedirs(dir_, exist_ok=True)
print("Data size (MB):", ds.nbytes/mb)
for i in range(nfiles):
path = os.path.join(dir_, "{:2d}.nc".format(i))
ds.to_netcdf(path)
Here are the versions of the libraries that I used:
Tensorflow: 2.8.0
Xarray: 0.18.2
netCDF4: 1.5.8
I will use this to compare several different ways of opening NetCDF files in TensorFlow. As with most things in TensorFlow, there are at least 3-4 different ways to skin the cat of loading NetCDF files. TensorFlow’s preferred native data structure for an iterable dataset of samples is tf.data.Dataset. This data structure can be fed to keras models.
First, I define some ways to open NetCDF data:
# from generator method
def load_nc_dir_with_generator(dir_):
def gen():
for file in glob.glob(os.path.join(dir_, "*.nc")):
ds = xarray.open_dataset(file, engine='netcdf4')
yield {key: tf.convert_to_tensor(val) for key, val in ds.items()}
sample = next(iter(gen()))
return tf.data.Dataset.from_generator(
gen,
output_signature={
key: tf.TensorSpec(val.shape, dtype=val.dtype)
for key, val in sample.items()
}
)
def load_nc_dir_with_map_and_xarray(dir_):
def open_path(path_tensor: tf.Tensor):
ds = xarray.open_dataset(path_tensor.numpy().decode())
return tf.convert_to_tensor(ds["a"])
return tf.data.Dataset.list_files(os.path.join(dir_, "*.nc")).map(
lambda path: tf.py_function(open_path, [path], Tout=tf.float64),
)
As you can see, these routines call some non-TensorFlow functions before instantiating the dataset object. This custom logic will be harder to parallelize or train on e.g. TPUs. The first solution load_nc_with_generator is perhaps the simplest, we first define a generator that opens each NetCDF and converts its contents to a dictionary of tf.Tensor objects. Because the generator is sequential, we won’t be able to parallelize it easily, although we could potentially use tf.data.Dataset.prefetch to load results ahead of time. For this reason, I implemented a version load_nc_dir_with_map_and_xarray that uses tf.data.Dataset.map, which is a potentially parallelizable operation.
Both methods require specifying the “schema” of the data. By schema, I mean the names, dtypes, and shapes of the arrays contained in the NetCDF. This is a key challenge with using the tf.data.Dataset object, and one reason why TensorFlow’s documentation on tfRecords is so byzantine and hard to read. This is where a self-describing format like NetCDF shines. The schema can be inferred from the data itself as we do when creating the tf.TensorSpec object in load_nc_dir_with_generator. I was lazy, so I didn’t bother with this in load_nc_dir_with_map_and_xarray, and just hardcoded the name "a" and data type tf.float64 for the sake of simplicity.
In any case, NetCDF data is easy to get the schema for but is not native to TensorFlow. By native, I mean methods that are built into TensorFlow’s graph like the functions in tf.io. These are usually backed by fast C-code and can be deployed easily in a variety of contexts including on TPUs. We might expect native formats to outperform NetCDF.
To see if this is true, I also benchmark routines which load the following TensorFlow-native formats:
- tfrecord
- the format saved by
tf.data.experimental.save
The following two functions first transform the NetCDFs to the TensorFlow native format, saving the result to disk. Then, they open this data using standard TensorFlow I/O connectors.
def load_nc_dir_cached_to_tfrecord(dir_):
"""Save data to tfRecord, open it, and deserialize
Note that tfrecords are not that complicated! The simply store some
bytes, and you can serialize data into those bytes however you find
convenient. In this case, I serialie with `tf.io.serialize_tensor` and
deserialize with `tf.io.parse_tensor`. No need for `tf.train.Example` or any
of the other complexities mentioned in the official tutorial.
"""
generator_tfds = load_nc_dir_with_generator(dir_)
writer = tf.data.experimental.TFRecordWriter("local.tfrecord")
writer.write(generator_tfds.map(lambda x: tf.io.serialize_tensor(x["a"])))
return tf.data.TFRecordDataset("local.tfrecord").map(
lambda x: tf.io.parse_tensor(x, tf.float64))
def load_nc_dir_after_save(dir_):
generator_tfds = load_nc_dir_with_generator(dir_)
tf.data.experimental.save(generator_tfds, "local_ds")
return tf.data.experimental.load("local_ds")
If there weren’t enough ways to save tf.data.Datasets to disk, they also have a .cache feature. This can either cache the data to disk or memory. Of course, the on-disk format saved by .cache is completely undocumented, but maybe that’s a good thing given my experience with TensorFlow documentation 😉. Here are two functions that cache to disk and memory:
def load_nc_dir_cache_to_disk(dir_):
generator_tfds = load_nc_dir_with_generator(dir_)
cached = generator_tfds.cache(f"{dir_}/.cache")
list(cached)
return cached
def load_nc_dir_cache_to_mem(dir_):
generator_tfds = load_nc_dir_with_generator(dir_)
cached = generator_tfds.cache()
list(cached)
return cached
Finally, we compare all the above against a method that simply reads the raw bytes from the NetCDF files into memory. This is an upper bound on the speed of data loading since it has no serialization/de-serialization overhead.
def load_nc_dir_to_bytes(dir_):
return tf.data.Dataset.list_files(os.path.join(dir_, "*.nc")).map(tf.io.read_file)
In summary, the following function lists all the methods we will benchmark:
def get_datasets(dir_):
return dict(
generator = load_nc_dir_with_generator(dir_),
map = load_nc_dir_with_map_and_xarray(dir_),
tfrecord = load_nc_dir_cached_to_tfrecord(dir_),
tf_data_save = load_nc_dir_after_save(dir_),
cache_disk = load_nc_dir_cache_to_disk(dir_),
bytes_only = load_nc_dir_to_bytes(dir_),
cache_mem = load_nc_dir_cache_to_mem(dir_),
)
Now, I will benchmark the datasets returned by these functions. The benchmark is simple: load the entire multi-file dataset twice. This is done by calling list(dataset). These benchmarks are performed for file sizes ranging from 256 kb to 8 mb. The total size of the dataset is 128 mb.
import timeit
total_size = 128 * mb
def show_timings(dir_):
datasets = get_datasets(dir_)
return {
key: timeit.timeit(lambda: list(datasets[key]), number=2)
for key in datasets
}
kb_range = [256, 1024, 4000, 8000]
for kb in kb_range:
save_dirs(kb * 1024, total_size, f"{dir_}/{kb}")
timings = {}
for kb in kb_range:
print(f"Working on {kb}")
timings[kb] = show_timings(f"{dir_}/{kb}")
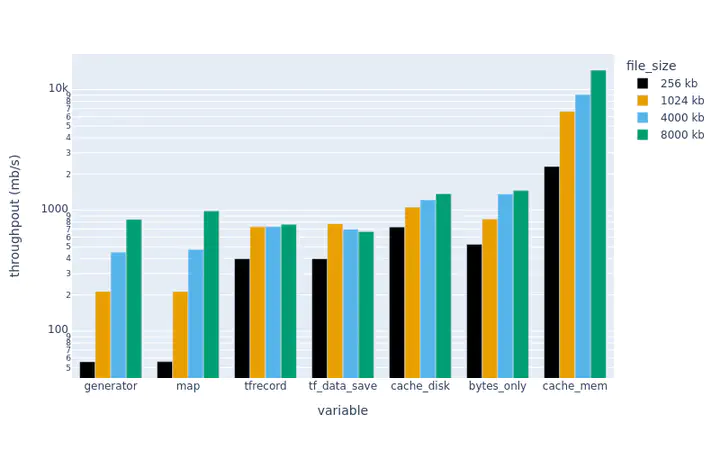
Finally, here are the results:
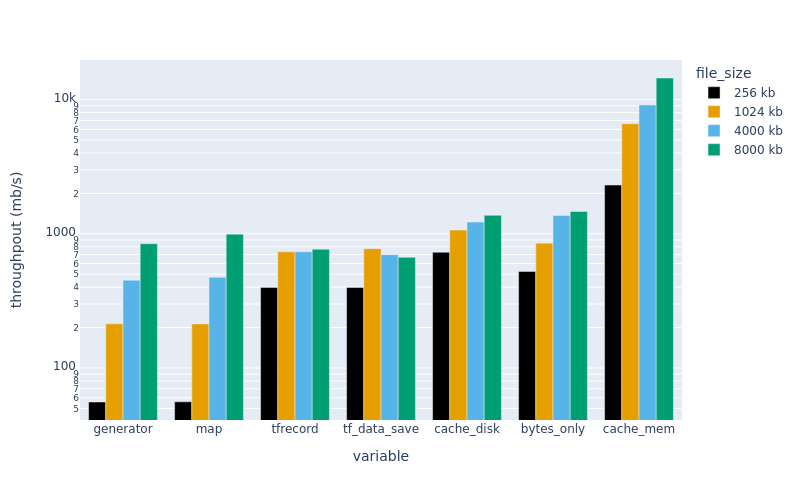
The plots above show throughput (higher is better).
We can see that the methods with writing custom loaders (map, generator) are very slow for small file sizes…but competitive for larger sizes. These examples were serial…so the TensorFlow native formats (tf_data_save and tfrecord) may become more competitive with proper parallelization.
The bytes-only dataset is a good comparison. It is an upper bound on how fast data can be read from a given file…suggesting that the NetCDF-reading approaches (map and generator) lose time converting bytes to tensors rather than reading bytes from disk. The serialization overhead is large for small files.
Preloading a dataset in memory (cache_mem) greatly increases the throughput….but caching on disk is fast too, and only requires one extra line of code. In practice, we have found that using .cache can lead to hard-to-debug memory leaks when training certain architectures, so YMMV.
Speed of deserializing a record
Much of the overhead of opening small NetCDFs comes not from loading from disk, but from decoding the loaded bytes.
Just how slow is this process? This code loads the raw bytes from a single 256 kb and times how long it takes to decode into a tensor:
read_ds = load_nc_dir_to_bytes("ncfiles/256")
item = next(iter(read_ds))
def open_bytes(bytes_tensor: tf.Tensor):
nc_bytes = bytes_tensor.numpy()
# syntax from https://unidata.github.io/netcdf4-python/
d = nc.Dataset('dummy.nc', memory=nc_bytes)
return tf.convert_to_tensor(d["a"][:])
%timeit open_bytes(item)
1000 loops, best of 5: 2.15 ms per loop
For this small amount of data, this process is MUCH slower than TensorFlow’s builtin tf.io.parse_tensor routine:
# is tensorflow io tensor serialization faster?
ds = open_bytes(item)
bytes_tensor = tf.io.serialize_tensor(tf.convert_to_tensor(ds))
%timeit tf.io.parse_tensor(bytes_tensor, tf.float64)
The slowest run took 5.32 times longer than the fastest. This could mean that an intermediate result is being cached.
10000 loops, best of 5: 82.3 µs per loop
So basically, opening many tiny NetCDF files will be quite slow.
Conclusions
In conclusion, NetCDF is a perfectly suitable format for feeding a TensorFlow training pipeline. It is self-describing and performs as well or better than “TensorFlow-native” formats like tfRecords or tf.data.experimental.save provided the data is chunked into NetCDF files containing at least a few MB of data. In practice, I have easily saturated a GPU when training from NetCDF data. At the same time, I can open the data and analyze it using familiar tools, and I could easily share it with colleagues.
Note that all the benchmarks here were single-threaded, and didn’t explore some of the more advanced tf.data.Dataset features such as parallel data loading or prefetching.
Moreover, because NetCDF I/O connectors require custom python code and dependencies, they cannot be as easily used in many standard cloud ML environments and cannot be integrated with more exotic performance optimizations like running on TPUs.
If you value these things, then it may make sense to roll your own custom serialization/deserialization logic and save the result to tfRecords.
I plan to show how to do this with TensorFlow datasets in a future post.